Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations - PubMed (original[)) (raw)
Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations
Olga Raskina et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Data are presented on quantum speciation in the Sitopsis section of the genus Aegilops (Poaceae, Monocotyledones). Two small, peripheral, isolated, wild populations of annual cross-pollinated Ae. speltoides and annual self-pollinated Ae. sharonensis are located 30 m apart on different soil types. Despite the close proximity of the two species and their close relatedness, no mixed groups are known. Comparative molecular cytogenetic analysis based on the intrapopulation variability of rRNA-encoding DNA (rDNA) chromosomal patterns of individual Ae. speltoides geno-types revealed an ongoing dynamic process of permanent chromosomal rearrangements. Chromosomal mutations can arise de novo and can be eliminated. Analysis of the progeny of the investigated genotypes testifies that inheritance of de novo rDNA sites happens frequently. Heterologous recombination and/or transposable elements-mediated rDNA transfer seem to be the mechanisms for observed chromosomal repatterning. Consequently, several modified genomic forms, intermediate between Ae. speltoides and Ae. sharonensis, permanently arise in the studied wild population of Ae. speltoides, which make it possible to recognize Ae. sharonensis as a derivative species of Ae. speltoides, as well as to propose rapidness and canalization of quantum speciation in Sitopsis species.
Figures
Fig. 1.
Geographical location and photos of the investigated populations. (a) Satellite image of eastern Mediterranean. (b) Field position of the studied populations. (c) Ae. sharonensis. (d) Ae. speltoides. The photographs of Ae. sharonensis and Ae. speltoides were taken in the same day. Although Ae. speltoides is blossoming, Ae. sharonensis is already mature.
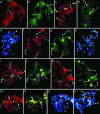
Fig. 2.
In situ hybridization of 5S (detected red) and 45S rDNA (detected green) on metaphase chromosomes of Aegilops speltoides and Ae. sharonensis and differential staining with Hoechst 33258. (a and b) Ae. speltoides Ts-24, standard karyotype. (c_–_e) Ae. speltoides (Kishon), genotype 4-1. Original genotypes presented in (f_–_h) and their progeny (k and l) carry additional 5S and/or 45S rDNA sites on chromosomes 1, 5, and 6. (f_–_h) Genotype 1-1, heterozygote for deletion of chromosome 1 long arm. (i and j) Genotype 1-2, from the same spike as genotype 1-1. (k and l) Progeny of the genotype 1-1 after self-pollination, homozygote for deletion of chromosome 1 long arm. (_m_–o) Ae. sharonensis (Kishon). Chromosomes 5 and 6 carry additional rDNA sites. (p) Ae. sharonensis (Caesarea), standard karyotype. Chromosomes 1, 5, and 6 are shown by blue, yellow, and white arrows, respectively. B-chromosomes are shown by white arrowheads.
Fig. 3.
Morphological and karyotypic variability of selected genotypes of Aegilops speltoides and Ae. sharonensis. Spikes of different genotypes are presented. Ae. speltoides ssp. aucheri Ts-84 and genotype 1: only terminal spikelet awned. Ae. speltoides ssp. ligustica Ts-24 and genotypes 4-1, 4-2, Ae. sharonensis: terminal spikelet and lemmas of lateral spikelets awned. Presence or absence of keel tooth on the glume is shown, respectively, by red and blue arrows in the enlargement fragments of the spikes (Ae. speltoides ssp. aucheri Ts-84 and genotype 4-1 do not have the tooth on the glume, red arrows). Simultaneous in situ hybridization of 5S (detected red) and 45S rDNA (detected green) on the somatic chromosomes. Only chromosomes 1, 5, and 6 with signals are shown. Polymorphism in the morphological characters and chromosomal rDNA patterns are observed in plants of Ae. speltoides and Ae. sharonensis from the Kishon.
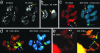
Fig. 4.
Intragenomic dynamics of rDNA sites in male gametogenesis of Ae. speltoides.(a) Intragenomic En/_Spm_-mediated transfers of 5S rDNA. Probe As5SDNAE (detected green) for detection of 5S rDNA and clone ESas-2 (detected red) for detection of the internal part of TPase region of En/_Spm_-like transposons were hybridized simultaneously on meiotic chromosomes. Extra 5S rDNA sites coupled to En/Spm transposons are shown by white arrows. (b) Chromosome 5, concurrence of En/Spm and additional 5S rDNA clusters are shown by yellow arrows. (c) Meiotic chromosomes (diakinesis stage) of the progeny of the same genotype after selfing. Chromosomes 1 and 6 carry additional 5S rDNA blocks concurrent with 45S rDNA. Moreover, one of the conjugated homologues of chromosome 1 carries one more additional intercalary 5S rDNA block on the short arm. One of the conjugated homologues of chromosome 5 carries an extra 5S rDNA block (3 blocks on bivalent totally). Chromosome 6 demonstrates a reduced satellite. (d) Meiotic chromosomes from the same anther as c. The consequences of heterologous recombination are observed. The bridge between 45S rDNA blocks of the chromosomes 1 and 6 still exist. Third (additional) 5S rDNA block on chromosome 5 is lost, whereas the other bivalent (green arrow) simultaneously carries 5S, 45S rDNA blocks and telomeric repeats (detected blue). (e) Meiotic chromosomes, diplotene stage. Heterologous synapses of chromosomes 1 and 5. Chromosome 5 carries an extra 45S rDNA block. Chromosomes 1, 5, and 6 are shown by blue, yellow, and white arrows, respectively. B-chromosomes are shown by white arrowheads.
Similar articles
- Diversity of long terminal repeat retrotransposon genome distribution in natural populations of the wild diploid wheat Aegilops speltoides.
Hosid E, Brodsky L, Kalendar R, Raskina O, Belyayev A. Hosid E, et al. Genetics. 2012 Jan;190(1):263-74. doi: 10.1534/genetics.111.134643. Epub 2011 Oct 31. Genetics. 2012. PMID: 22042572 Free PMC article. - [Molecular analysis of the phylogenetic relationships among the diploid aegilops species of the section Sitopsis].
Goriunova SV, Chikida NN, Kochieva EZ. Goriunova SV, et al. Genetika. 2008 Jan;44(1):137-41. Genetika. 2008. PMID: 18409395 Russian. - Fate of Aegilops speltoides-derived, repetitive DNA sequences in diploid Aegilops species, wheat-Aegilops amphiploids and derived chromosome addition lines.
Kumar S, Friebe B, Gill BS. Kumar S, et al. Cytogenet Genome Res. 2010 Jul;129(1-3):47-54. doi: 10.1159/000314552. Epub 2010 Jun 15. Cytogenet Genome Res. 2010. PMID: 20551615 - Gametocidal genes of Aegilops: segregation distorters in wheat-Aegilops wide hybridization.
Niranjana M. Niranjana M. Genome. 2017 Aug;60(8):639-647. doi: 10.1139/gen-2017-0023. Epub 2017 Jun 27. Genome. 2017. PMID: 28654760 Review. - Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes.
Raskina O, Barber JC, Nevo E, Belyayev A. Raskina O, et al. Cytogenet Genome Res. 2008;120(3-4):351-7. doi: 10.1159/000121084. Epub 2008 May 23. Cytogenet Genome Res. 2008. PMID: 18504364 Review.
Cited by
- Chromosomal Localization and Diversity Analysis of 5S and 18S Ribosomal DNA in 13 Species from the Genus Ipomoea.
Wu J, Lang T, Zhang C, Yang F, Yang F, Qu H, Pu Z, Feng J. Wu J, et al. Genes (Basel). 2024 Oct 19;15(10):1340. doi: 10.3390/genes15101340. Genes (Basel). 2024. PMID: 39457464 Free PMC article. - Cytomolecular diversity among Vigna Savi (Leguminosae) subgenera.
Dias S, Souza RC, Vasconcelos EV, Vasconcelos S, da Silva Oliveira AR, do Vale Martins L, de Oliveira Bustamante F, da Costa VA, Souza G, da Costa AF, Benko-Iseppon AM, Knytl M, Brasileiro-Vidal AC. Dias S, et al. Protoplasma. 2024 Sep;261(5):859-875. doi: 10.1007/s00709-024-01944-z. Epub 2024 Mar 11. Protoplasma. 2024. PMID: 38467939 - Physical localization of 45S rDNA in Cymbopogon and the analysis of differential distribution of rDNA in homologous chromosomes of Cymbopogon winterianus.
Thakur S, Kumar U, Malik R, Bisht D, Balyan P, Mir RR, Kumar S. Thakur S, et al. PLoS One. 2021 Nov 18;16(11):e0257115. doi: 10.1371/journal.pone.0257115. eCollection 2021. PLoS One. 2021. PMID: 34793445 Free PMC article. - Full-length LTR retroelements in Capsicum annuum revealed a few species-specific family bursts with insertional preferences.
Yañez-Santos AM, Paz RC, Paz-Sepúlveda PB, Urdampilleta JD. Yañez-Santos AM, et al. Chromosome Res. 2021 Dec;29(3-4):261-284. doi: 10.1007/s10577-021-09663-4. Epub 2021 Jun 4. Chromosome Res. 2021. PMID: 34086192 - Molecular Cytogenetics of Eurasian Species of the Genus Hedysarum L. (Fabaceae).
Yurkevich OY, Samatadze TE, Selyutina IY, Romashkina SI, Zoshchuk SA, Amosova AV, Muravenko OV. Yurkevich OY, et al. Plants (Basel). 2021 Jan 4;10(1):89. doi: 10.3390/plants10010089. Plants (Basel). 2021. PMID: 33406686 Free PMC article.
References
- Timofeeff-Ressovsky, N. W., Vorontsov, N. N. & Yablokov, A. V. (1977) Evolutionary Theory Essay (Nauka, Moscow).
- White, M. J. D. (1978) Modes of Speciation (Freeman, San Francisco).
- Grant, V. (1981) Plant Speciation (Columbia Univ. Press, New York).
- Nevo, E. (2000) in Nature Encyclopedia of Life Sciences (Nature Publishing Group, London), pp. 1–11.
- Forsdyke, D. R. (2003) J. Biol. Sys. 11, 341–350.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources